Linear Models

# Prepare 3 clusters of data
I, J = (1000,2) # 1000 points, 2 attributes
np.random.seed (9527)
X = np.random.randn (I, J)
Mu0 = np.array([[0,-4], [-4,4], [2,2]]) # Centers of the 3 clusters
X += Mu0[np.random.choice(np.arange(3), I),:] # Assign points to the 3 clusters
# plt.scatter(X[:,0], X[:,1]) # Plot original data
K = Mu0.shape[0] # Define the number of clusters to generate (e.g., 3)
max_iter = 4 # Number of KNN iterations
colors = np.array(["C0", "C1", "C2"])
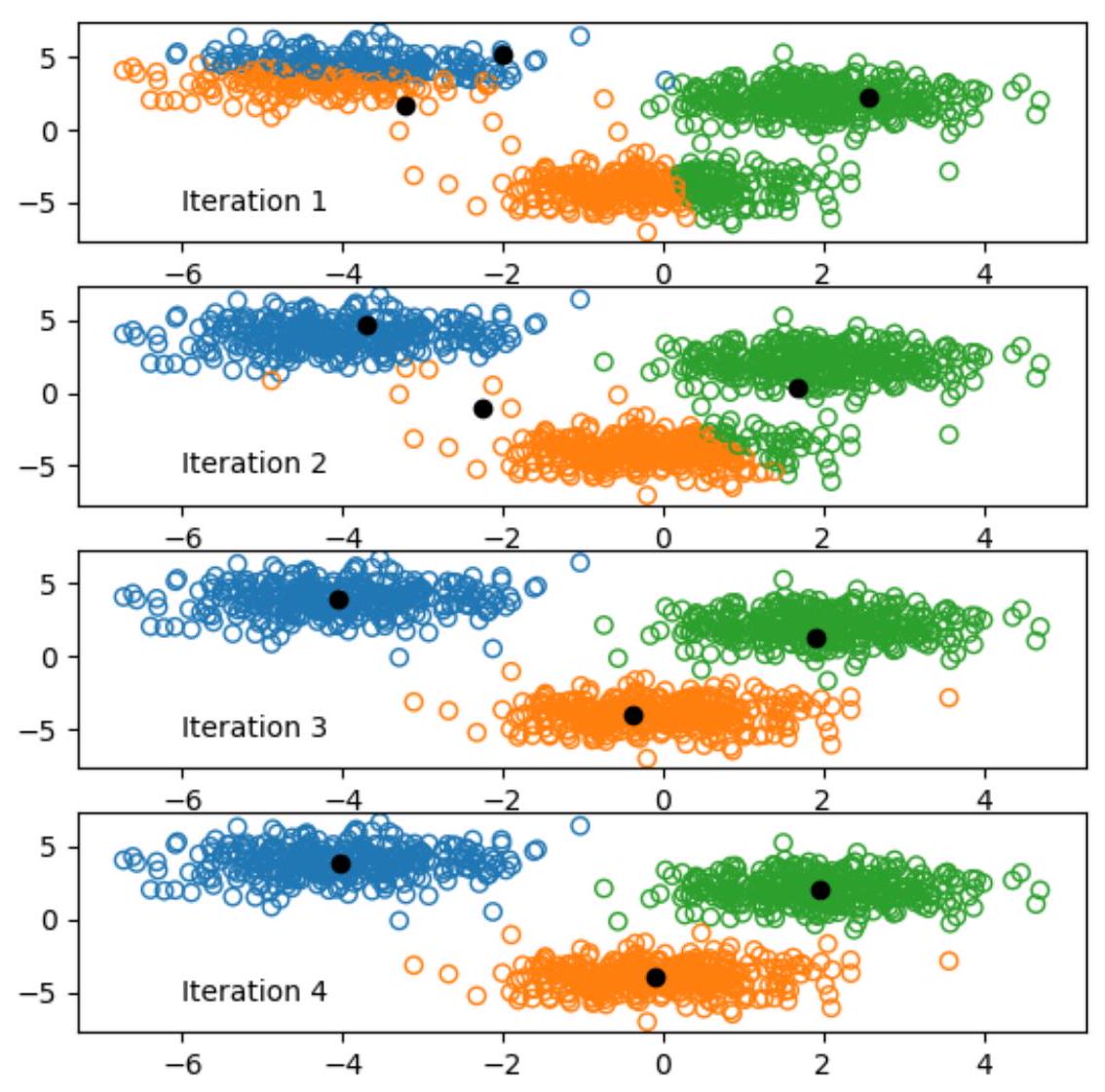
f,ax = plt.subplots(max_iter) # figsize=(6.0, 4.6*max_iter)
Mu = X[np.random.choice(X.shape[0],K),:]
for n in range(max_iter):
D = np.empty((X.shape[0],K)) # 1000 points, each has 3 distances to 3 centers
for i in range(K): # Loop over all the centers (3 centers)
D[:,i] = np.sum((X-Mu[i])**2,axis=1)
y = np.argmin(D,axis=1)
ax[n].scatter(X[:,0], X[:,1], edgecolors=colors[y], facecolors='none')
ax[n].legend(['Iteration '+str(n+1)],frameon=False, scatterpoints=0)
ax[n].scatter(Mu[:,0], Mu[:,1], c='k')
Mu = np.array([np.mean(X[y==k],axis=0) for k in range(K)])
loss = np.linalg.norm(X - Mu[np.argmin(D,axis=1),:])**2/X.shape[0]